DFG Heisenberg grant LE 3358/4-1 “DiaLReDox – the regulation of the photosynthetic light reaction, light dependent retrograde signaling and redox dynamics in the diatom Phaeodactylum tricornutum”
and DFG research grant LE3358/5-1 “Identifying the molecular transducers of retrograde redox signaling and unravelling compartmental redox dynamics in the diatom Phaeodactylum tricornutum”
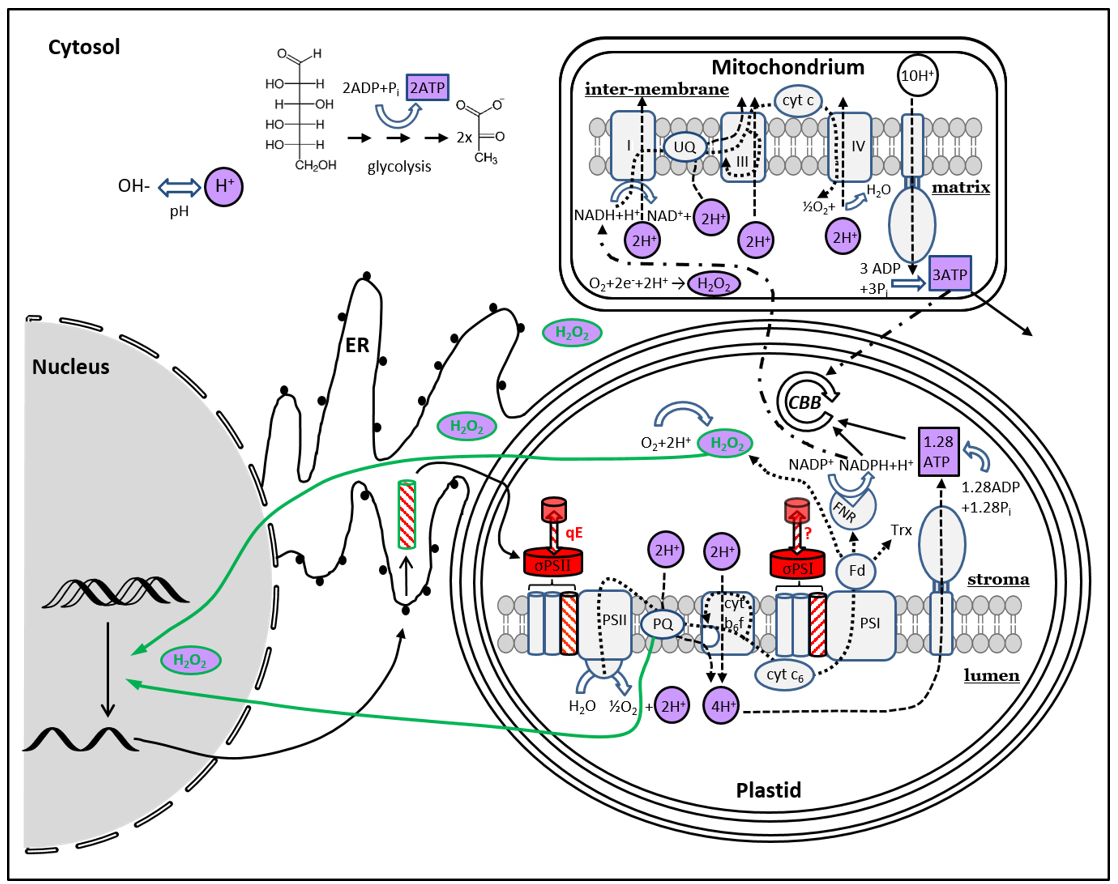
Diatoms have complex plastids, which they have obtained from a red alga via secondary endosymbiosis. Therefore, their plastid envelope consists of four membranes, compared to two in green algae and vascular plants. More than ten years ago, we demonstrated the existence of light-dependent retrograde redox signalling - the control of nuclear gene expression via a stimulus from the plastid - in the diatom Phaeodactylum tricornutum. We identified the redox state of the plastoquinone pool as the starting point of this signaling cascade. This is only one of many dynamically and light-dependent changing redox components in the diatom cell. For example, pH, ATP and H2O2 are dependent on the light conditions and are potentially influenced by retrograde signaling or control it themselves. Their compartmental dynamics are completely unknown in diatoms.
In this project, we aim to unveil molecular components of retrograde redox signaling in diatoms and gain insights into the signaling cascade that originates in the plastid and terminates in the nucleus. We are also interested in the light-dependent compartmental redox dynamics of pH, ATP and H2O2, especially with regard to the interplay of the photosynthetic light reaction with mitochondrial respiration.
Methodologically, we use transcriptome analyses, yeast-1-hybrid assays, fluorescent protein-based redox sensors, fluorescence microscopy and spectroscopy, and genome editing.
Together with the dynamic regulation of the light response, which is being investigated in the DFG project LE3358/3-2, the results of this project will enable a more holistic understanding of photophysiology. This will make it possible to establish new models for light acclimation in diatoms, but also in other taxa.
DFG research grant LE3358/3-2 “The molecular basics of rapid photoprotection and its influence on competitive fitness in Phaeodactylum tricornutum”
Diatoms are very successful in almost all aquatic habitats. One of the main reasons for this is assumed to be their pronounced photoprotective capacity based on qE (energy dependent fluorescence quenching). While the importance of the carotenoid diatoxanthin, which is formed in the xanthophyll cycle during exposure to strong light, for qE has long been known, the relevance of Lhcx proteins for qE was only revealed a few years ago. We here could show that Lhcx1, x2 and x3, but not Lhcx4, mediate qE in Phaeodactylum tricornutum.
In this project, we want to investigate the qE mechanism in more detail with regard to its molecular components. We are interested in the amino acids and peptide motifs that are essential to functionally distinguish an Lhcx protein from a classical light-harvesting protein. In addition, we want to identify interaction partners of the Lhcx proteins in the thylakoid membrane, because so far there are only clues as to where exactly this mechanism of qE takes place. Finally, we perform extensive growth experiments under varying stress conditions with different Lhcx mutants to reveal the actual influence of qEs under near-natural simulated conditions.
Methodologically, we use reverse genetics and genome editing, different biochemical approaches and a fleet of photophysiological devices.
Our results are integrated into the overall goal of the Heisenberg programme, the high-resolution and detailed understanding of the molecular and physiological response to light stress in diatoms.
